How terrible is malaria? (Data visualization with Python)
Malaria is a serious and sometimes fatal disease caused by a parasite that commonly infects a certain type of mosquito which feeds on humans. People who get malaria are typically very sick with high fevers, shaking chills, and flu-like illness. Because malaria causes so much illness and death, the disease is a great drain on many national economies. Since many countries with malaria are already among the poorer nations, the disease maintains a vicious cycle of disease and poverty.
In this article, I would share 3 visualizations about Malaria using Python. You can find my Github repository for this blog here. Data source: https://github.com/rfordatascience/tidytuesday/tree/master/data/2018/2018-11-13.
1. The spread of malaria worldwide over time.
I hope to have an overall understanding of the spread of malaria in the world, so I created an interactive graph to show the spread of deaths from malaria over time. To do this, I preprocessed the malaria death data and downloaded the map data from here. Then I used TimeSliderChoropleth in the Folium package to make a choropleth with a timeslider, to show the spread of malaria over time.
import branca.colormap as cm
import folium
import geopandas as gpd
from folium.plugins import TimeSliderChoropleth
## load data
death = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_deaths.csv')
death_age = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_deaths_age.csv')
inc = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_inc.csv')
countries = gpd.read_file('Countries_WGS84.shp')
countries = countries.rename(columns={'CNTRY_NAME': 'Entity'})
## data preprocessing
death.columns = ['death_rate' if x=='Deaths - Malaria - Sex: Both - Age: Age-standardized (Rate) (per 100,000 people)' else x for x in death.columns]
sorted_df = death.sort_values(['Entity',
'Year']).reset_index(drop=True)
sorted_df['date'] = sorted_df['Year'].apply(lambda x: datetime.datetime(x+1, 1, 1))
sorted_df['date'] = pd.to_datetime(sorted_df['date'], yearfirst=True).astype(int) / 10**9
sorted_df['date'] = sorted_df['date'].astype(int)
joined_df = sorted_df.merge(countries, on='Entity')
joined_df = joined_df[['Entity', 'death_rate', 'date', 'geometry']]
## create the map
max_colour = max(joined_df['death_rate'])
min_colour = min(joined_df['death_rate'])
cmap = cm.linear.YlOrRd_09.scale(min_colour, max_colour)
joined_df['colour'] = joined_df['death_rate'].map(cmap)
country_list = joined_df['Entity'].unique().tolist()
country_idx = range(len(country_list))
countries_df = joined_df[['geometry']]
countries_gdf = gpd.GeoDataFrame(countries_df)
countries_gdf = countries_gdf.drop_duplicates().reset_index()
style_dict = {}
for i in country_idx:
country = country_list[i]
result = joined_df[joined_df['Entity'] == country]
inner_dict = {}
for _, r in result.iterrows():
inner_dict[r['date']] = {'color': r['colour'], 'opacity': 0.7}
style_dict[str(i)] = inner_dict
slider_map = folium.Map(min_zoom=2, max_bounds=True, tiles='cartodbpositron')
_ = TimeSliderChoropleth(
data=countries_gdf.to_json(),
styledict=style_dict,
).add_to(slider_map)
_ = cmap.add_to(slider_map)
cmap.caption = "Malaria Deaths (per 100,000 people)- Sex: Both - Age: Age-standardized"
slider_map.save(outfile='malaria_deaths.html')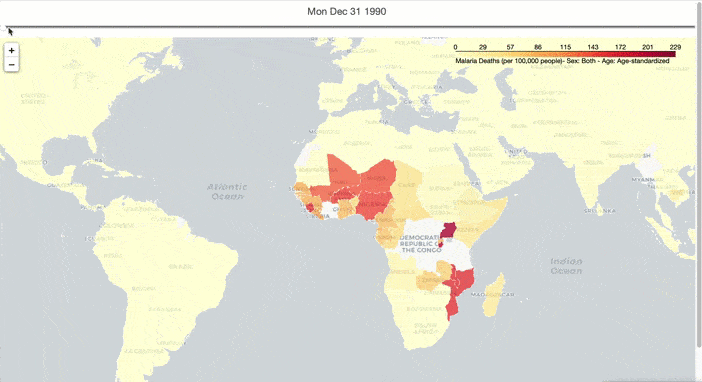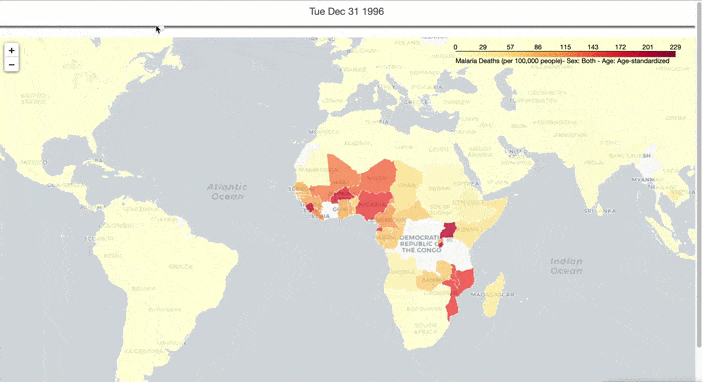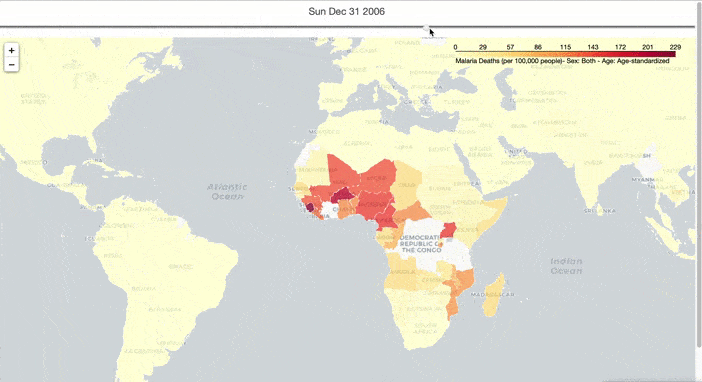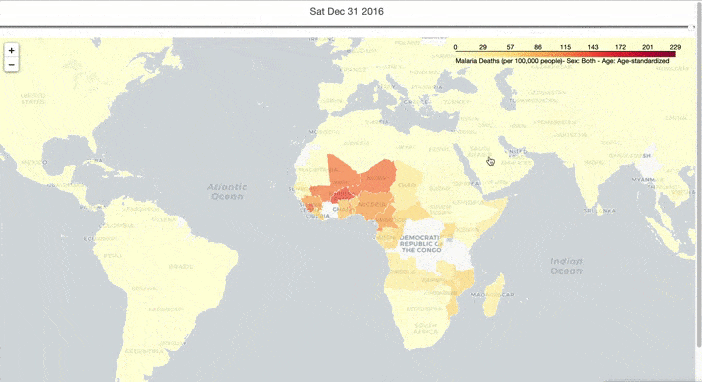
From Figure 1, the global spread of malaria has been curbed over time. However, Malaria occurs mostly in poor tropical and subtropical areas of the world, and Africa is still the most affected region due to a combination of factors:
- A very efficient mosquito (Anopheles gambiae complex) is responsible for high transmission.
- Local weather conditions often allow transmission to occur year round.
- Scarce resources and socio-economic instability have hindered efficient malaria control activities.
2. The relationship between malaria incidence and mortality.
How terrible is malaria? What will happens if you have malaria? How likely are you to have the worst result? Which countries do better at curbing the spread and controlling malaria? Let us discuss the relationship between the incidence and mortality of malaria. I linked malaria death dataset with the age dataset and create a scatter plot for the 20 most-affected countries among the fifteen years. Then I add a smooth line to each country. Figure 2 shows the changes in the incidence and mortality of malaria in the 20 most-affected countries.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import make_interp_spline
import matplotlib.lines as mlines
## load data
death = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_deaths.csv')
death_age = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_deaths_age.csv')
inc = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_inc.csv')
## tidy data
joined_df = inc.merge(death, on=('Entity', 'Year'), how='inner')
joined_df = joined_df.rename(columns = {'Incidence of malaria (per 1,000 population at risk) (per 1,000 population at risk)':'Incidence', 'Deaths - Malaria - Sex: Both - Age: Age-standardized (Rate) (per 100,000 people)': 'Deaths'})
countries = joined_df.groupby('Entity').min().nlargest(20, 'Incidence').index
## create plot
fig = plt.figure(figsize = (15, 10))
ax = fig.add_subplot(111)
markers = ['P', '^', 'o', '*']
colors = dict(zip(countries, plt.cm.jet(np.linspace(0, 1, len(countries)))))
for country, color in colors.items():
df = joined_df[joined_df['Entity'] == country]
for i in range(4):
ax.scatter(df['Incidence'].iloc[i], df['Deaths'].iloc[i], color = color, marker = markers[i], s = 100)
x = np.array(df['Incidence'])
y = np.array(df['Deaths'])
y_smooth = make_interp_spline([2000, 2005, 2010, 2015], np.c_[x, y])(np.linspace(2000, 2015, 100))
ax.plot(*y_smooth.T, color = color, label = country)
ax.set_xlabel('Incidence of Malaria (per 1,000 population at risk)', fontsize=20)
ax.set_ylabel('Death of Malaria (per 100,000 people)', fontsize=20)
ax.set_title('Incidence rate and death rate of Malaria for 20 most-affected countries', fontsize=20)
leg1 = ax.legend(handles=[
mlines.Line2D([], [], color='black', marker='P', linestyle='None', label='2000', markersize = 10),
mlines.Line2D([], [], color='black', marker='^', linestyle='None', label='2005', markersize = 10),
mlines.Line2D([], [], color='black', marker='o', linestyle='None', label='2010', markersize = 10),
mlines.Line2D([], [], color='black', marker='*', linestyle='None', label='2015', markersize = 10)],
prop={'size': 15})
leg2 = ax.legend(loc='upper left', bbox_to_anchor=(1.02, 1), borderaxespad=0, prop={'size': 15})
ax.add_artist(leg1)
pass
From Figure 2, we can see that morbidity and mortality in most countries have declined over time, but in some countries, such as Sierra Leone, Equatorial Guinea, Burkina Faso and Malawi, malaria has worsened for several years and then eased. It is worth moting that morbidity and mortality rates in the Gambia have decreased rapidly over time, showing that Gambia has played an excellent role in controlling the spread of malaria and treating the disease.
3. The age structure of malaria mortality.
Let us study the death of malaria in more depth. What is the age structure of malaria deaths? Which age group has the highest risk of malaria? Similarly, I create a scatter plot on the age structure of deaths from malaria for the 20 most-affected countries. Figure 3 shows the age structure of malaria deaths in the 20 most-affected countries.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import make_interp_spline
## load data
death = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_deaths.csv')
death_age = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_deaths_age.csv')
inc = pd.read_csv('tidytuesday/data/2018/2018-11-13/malaria_inc.csv')
# tidy death_age
## remove duplicate index column
death_age = death_age.drop(labels = 'Unnamed: 0', axis = 1)
## rename entity to Entity
death_age.columns = ['Entity' if x=='entity' else x for x in death_age.columns]
## generate an indicator order to sort df
death_age['age'] = death_age['age_group'].replace({'Under 5': 0, '5-14': 1, '15-49': 2, '50-69': 3, '70 or older': 4})
## remove entities that are not country
countries = list(np.unique(death_age['Entity']))
countries.remove('World')
countries.remove('Sub-Saharan Africa')
countries.remove('Western Sub-Saharan Africa')
countries.remove('Low SDI')
countries.remove('Low-middle SDI')
countries.remove('Eastern Sub-Saharan Africa')
countries.remove('Central Sub-Saharan Africa')
## create the plot and label the 3 most-affected countries
colors = plt.cm.jet(np.linspace(0, 1, len(countries)))
plt.figure(figsize=(16,10))
for i, country in enumerate(countries):
df = death_age[death_age['Entity'] == country]
df = df[df['year'] == 2016]
df = df.sort_values(['age']).reset_index(drop=True)
x = df['age']
y = df['deaths']
x_new = np.linspace(x.min(), x.max(), 300)
y_smooth = make_interp_spline(x, y)(x_new)
plt.scatter(x, y, c='black',alpha = 0.5)
## smooth line
plt.plot(x_new, y_smooth,c=colors[i], label=country)
plt.xticks(np.arange(min(x), max(x)+1, 1.0), ['Under 5', '5-14', '15-49', '50-69', '70 or older'])
plt.text(0.15, 175000, 'Nigeria')
plt.text(0.15, 50000, 'Democratic Republic of Congo')
plt.text(0.15, 27000, 'Niger')
plt.legend(loc='upper center', bbox_to_anchor=(0.5, -0.05), shadow=True, ncol=4)
plt.xlabel('age group')
plt.ylabel('deaths')
plt.title('Malaria Death among Different Age Groups in 2016 (latest)')
plt.show()
Children under 5 years of age are one of the most vulnerable groups affected by malaria, especially in Nigeria, the Democratic Republic of the Congo and Niger. In high transmission areas, partial immunity to the disease is acquired during childhood. In such settings, the majority of malarial disease, and particularly severe disease with rapid progression to death, occurs in young children without acquired immunity. Severe anaemia, hypoglycemia and cerebral malaria are features of severe malaria more commonly seen in children than in adults.
WHO recommends the following package of interventions for the prevention and treatment of malaria in children:
- use of long-lasting insecticidal nets;
- in areas with highly seasonal transmission of the Sahel sub-region of Africa, seasonal malaria chemoprevention (SMC) for children aged between 3 and 59 months;
- in areas of moderate-to-high transmission in sub-Saharan Africa, intermittent preventive therapy for infants, except in areas where WHO recommends administration of SMC;
- prompt diagnosis and effective treatment of malaria infections.
Based on the above analysis, we can conclude that although the spread of malaria has been suppressed to a certain extent, due to its serious consequences, we still need to work hard to control and find more effective treatments.